Welcome to Deep-PLA
Deep-PLA is a web server developed for understanding the reversible enzyme-specific lysine acetylation regulation for histone acetyltransferases (HATs) including CREBBP, EP300, HAT1, KAT2A, KAT2B, KAT5 and KAT8, and deacetylases (HDACs) including HDAC1, HDAC2, HDAC3, HDAC6, SIRT1, SIRT2, SIRT3, SIRT6, and SIRT7. Based on deep learning method, the predictor achieved promising performances. Four bright spots of Deep-PLA software are summarized below: i) four kinds of protein sequence features are modeled independently and then merged into one model; ii) particle swarm optimizer algorithm is used to optimize the hyperparameters; iii) the protein-protein interaction network and colocalization information are provided and visulized in the result page; iv) three fundamental properties of protein sequence are visualized in the result page, including disorder, surface accessibility and secondary structure.
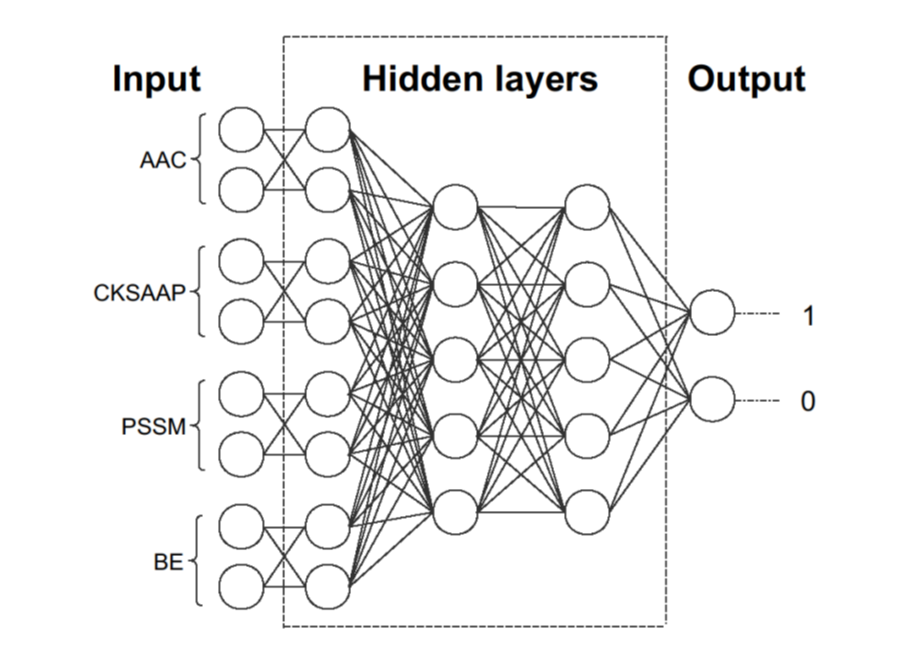
Deep neural network for prediction of HAT/HDAC-specific acetylation
• Four sequence features of the acetylation sites are extracted to build models.
• Four models are combined into one full connection layer.
• Deep-learning method is used to predict HAT/HDAC-specific acetylation sites.
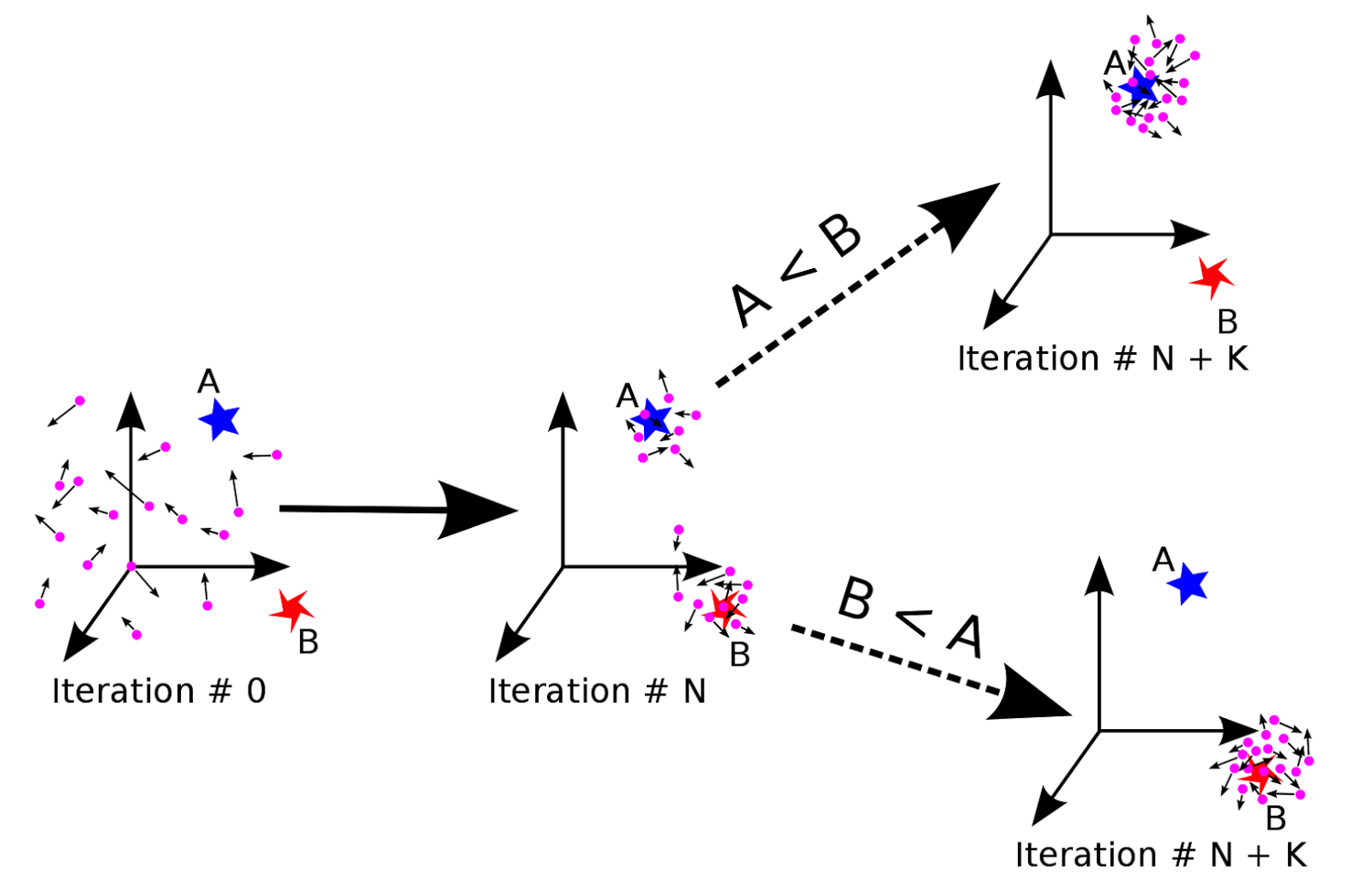
Optimize the hyperparameters of Deep-PLA
• Deep neural network contains hyperparameters such as learning rates, activation functions, dropout rates, etc.
• Taking both efficiency and effectiveness into account, particle swarm optimizer (PSO) is applyed to optimize the hyperparameters.
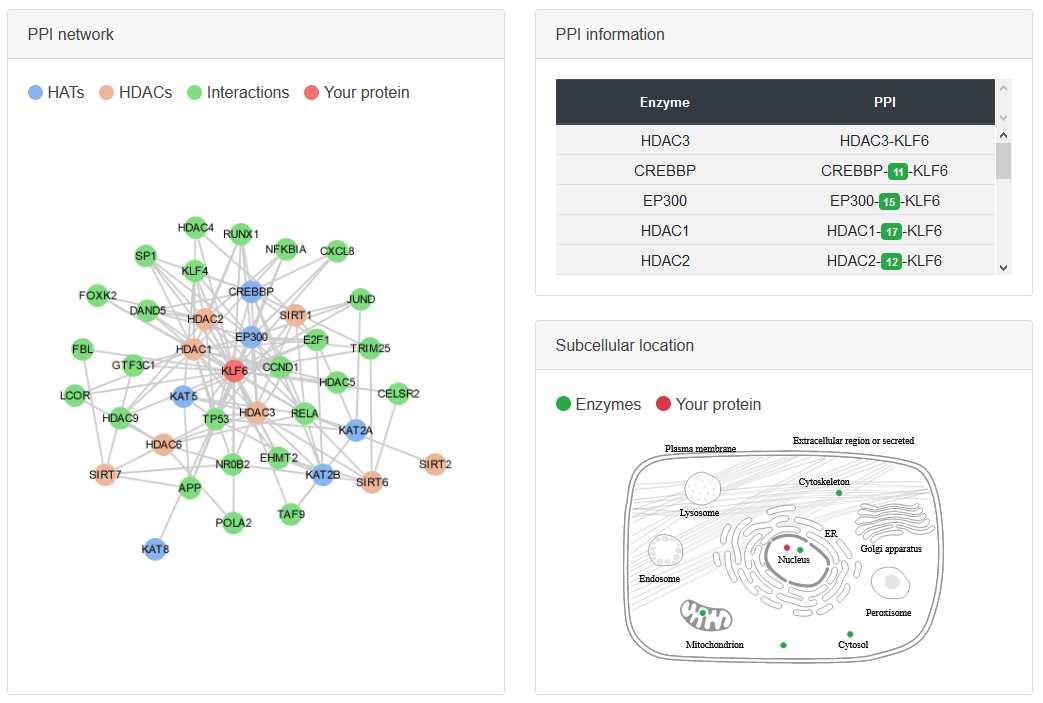
PPI network
• The protein-protein interaction (PPI) data are collected from several databases.
• The cell map shows the colocalization between the enzyme and the substrate.
• In addition to the direct PPI, the proteins connected to the enzymes via scaffold proteins are also shown.
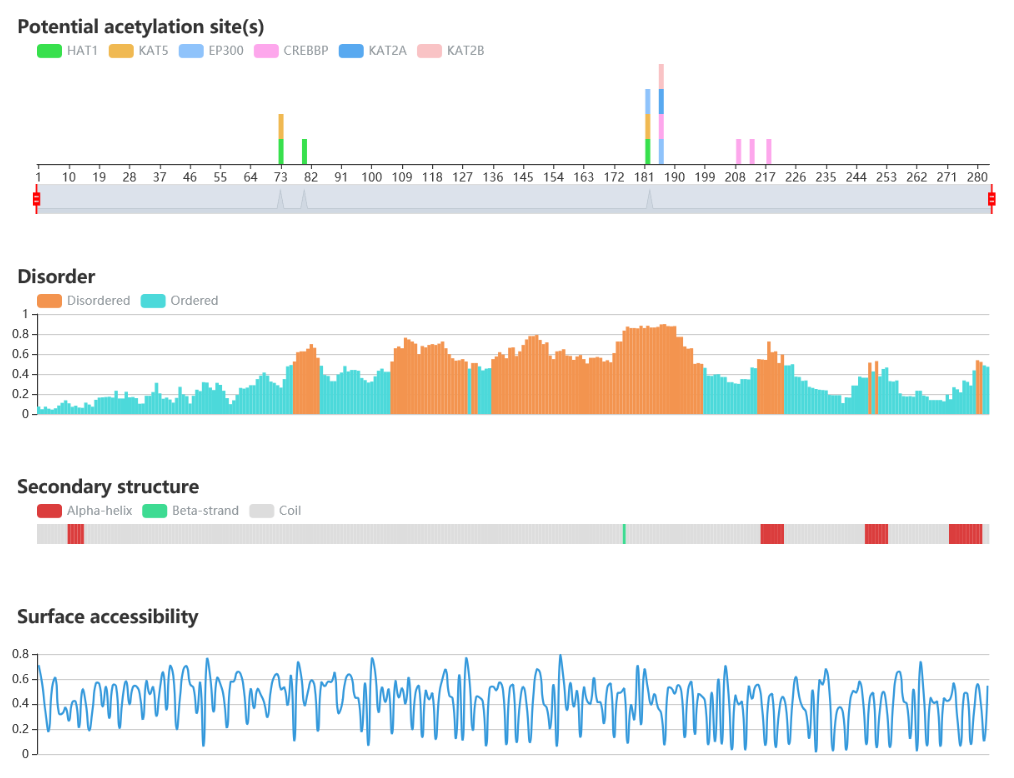
Protein sequence and structure properties
• IUPred is used to predict the disorder region.
• NetSurfP is used to predict the surface accessibility and secondary structure.
• The potential modified sites of the query sequence are indicated.
